Writing a SimPlot Clone in Python
SimPlot is a great little program written by Stuart Ray, have a look here.
As you’ve probably already gathered from the title, I’m going to go over how to recreate the basic functionality of SimPlot in Python. Why? I wanted to accomplish two things: 1. Figure out exactly what’s happening behind the scenes including how some edge cases are handled. 2. Have something which I can run easily on my Mac or Linux computer.
TL;DR Download the completed script from GitHub
How Does it Work?
Provided with an alignment of two or more sequences (nucleotide or amino acid), the original SimPlot will calculate the % identity between them within a rolling window and then plot it. See below.

Of course, in practice the window will usually be much larger, as will the sequences! First things first, let’s write some python to deal with the pointer and rolling window.
Creating the Pointer Class
We need to keep track of the current pointer position and the location of the window bounds (which we’ll call window_lower and window_upper) given the window size, step size, and current iteration. This is fairly simple but we need to remember that in Python all counters start at 0. We also need to decide on the pointer location — it always has to be an integer and so we can’t always just divide the window size in half.
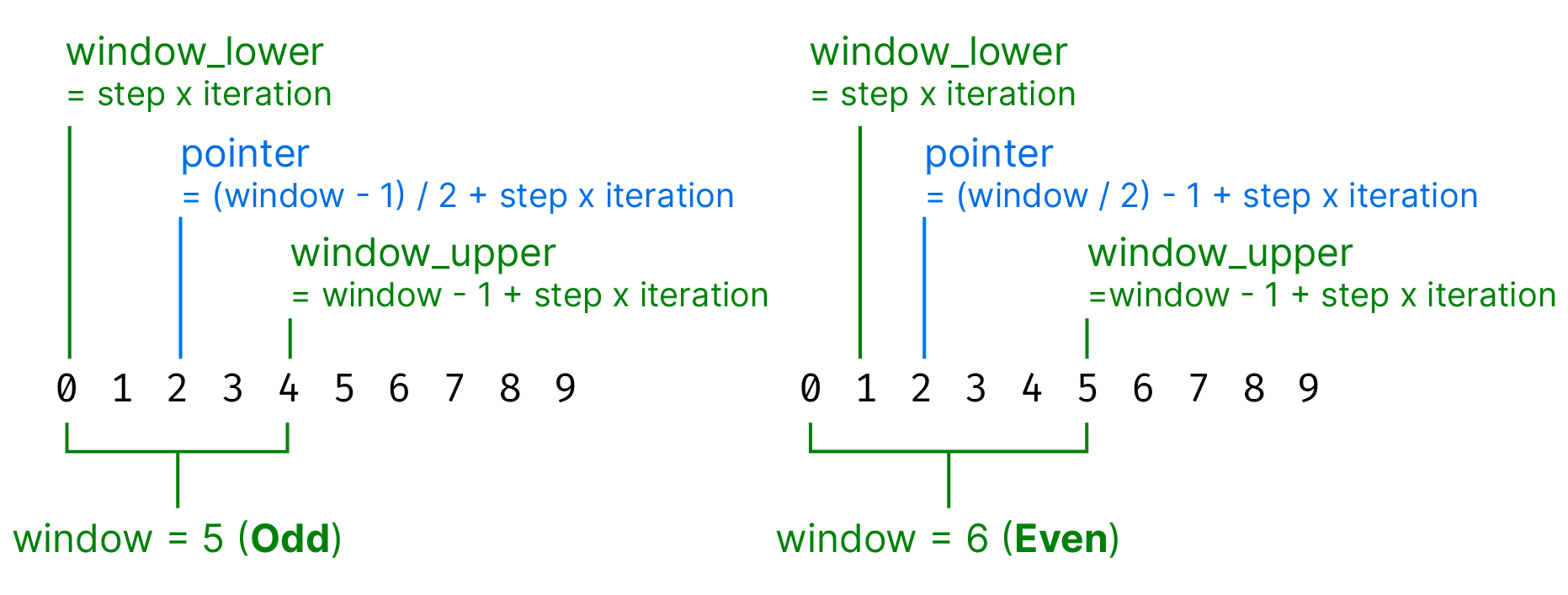
Looking at the image above you’ll notice that we need to know whether the the window size is even or odd to set the pointer position. For the window bounds the calculation is the same. The solution that I’ve come up with might be different to how you would have done it but that’s ok!
class Pointer:
# Set all initial values using calculate() on load
def __init__(self, window, step):
self.window = window
self.step = step
self.iteration = 0
self.calculate()
# Read the current parameters and set values
def calculate(self):
self.window_lower = self.step * self.iteration
self.window_upper = self.window - 1 + self.step * self.iteration
# Is the window size odd or even?
# Set correct pointer location
if (self.window % 2) == 0:
self.pointer = (self.window / 2) - 1 + self.step * self.iteration
else:
self.pointer = (self.window - 1) / 2 + self.step * self.iteration
# Increase the iteration count and recalculate all values
def increment(self):
self.iteration += 1
self.calculate()
# Reset the iteration count to zero and recalculate all values
def reset(self):
self.iteration = 0
self.calculate()Calculating Similarity
Using the positions provided by the Pointer class, we can now extract the sequences within the rolling window (from the full-length sequences) and compare them. To keep things simple, we’ll abstract this into two functions, extract and compare.
# Take the Pointer object and extract a windowed sequence
# from the full-length sequence
def extract(pointer, sequence):
# Get the window boundaries
lower = pointer.window_lower
upper = pointer.window_upper
windowed_sequence = sequence[lower:upper]
return windowed_sequenceThe extract function is not super neccessary (it can easily be shortened to one line) but it’s useful to have this functionality separate in case we build upon this in future.
We can run extract twice — once per sequence — and then pass the outputs into the compare function to get the identity.
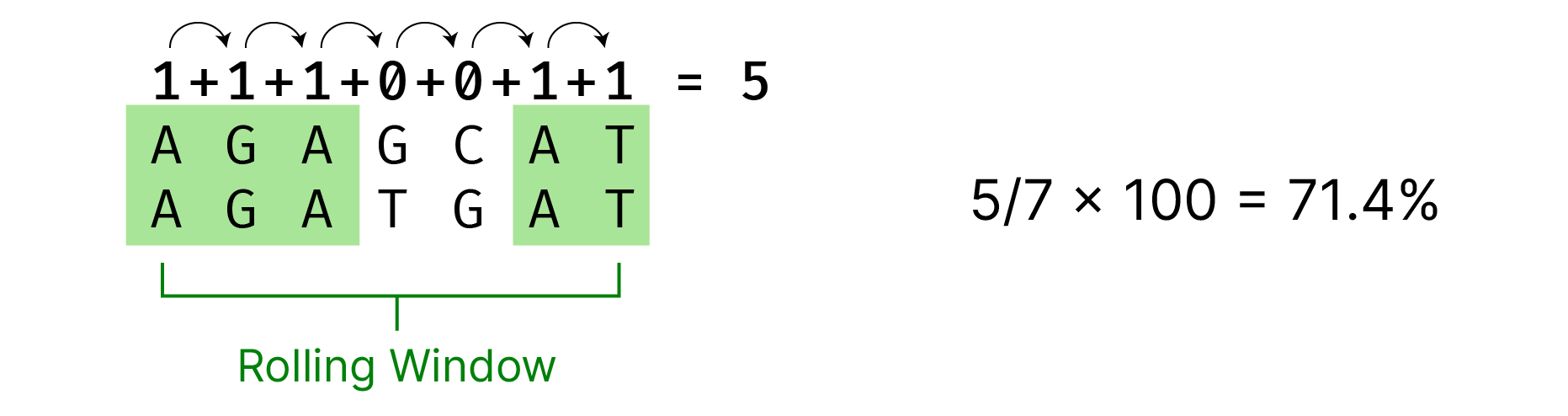
def compare(sequenceA, sequenceB):
length = len(sequenceA)
# Initiate a counter
sum = 0
# Look at each position in turn
for base in range(0, length):
# Is the base/residue the same?
if ( sequenceA[base] == sequenceB[base] ):
# Increase the counter
sum += 1
# Convert the final count to a percentage
identity = sum / length * 100
return identityNow we pretty much have all the basics sorted, lets create the main loop.
The Main Loop
We need a way to get the aligned sequences into the script, for this tutorial we’ll just paste them in as strings but for the final program I’ll write a quick class to extract them from a file.
sequenceA = "AGAGCATGGATGGCAAGAAAGTT----ATAA ...continued "
sequenceB = "AGATGATGGATGGGGAGAAAGATCACTATAA ...continued "Now to put everything above together…
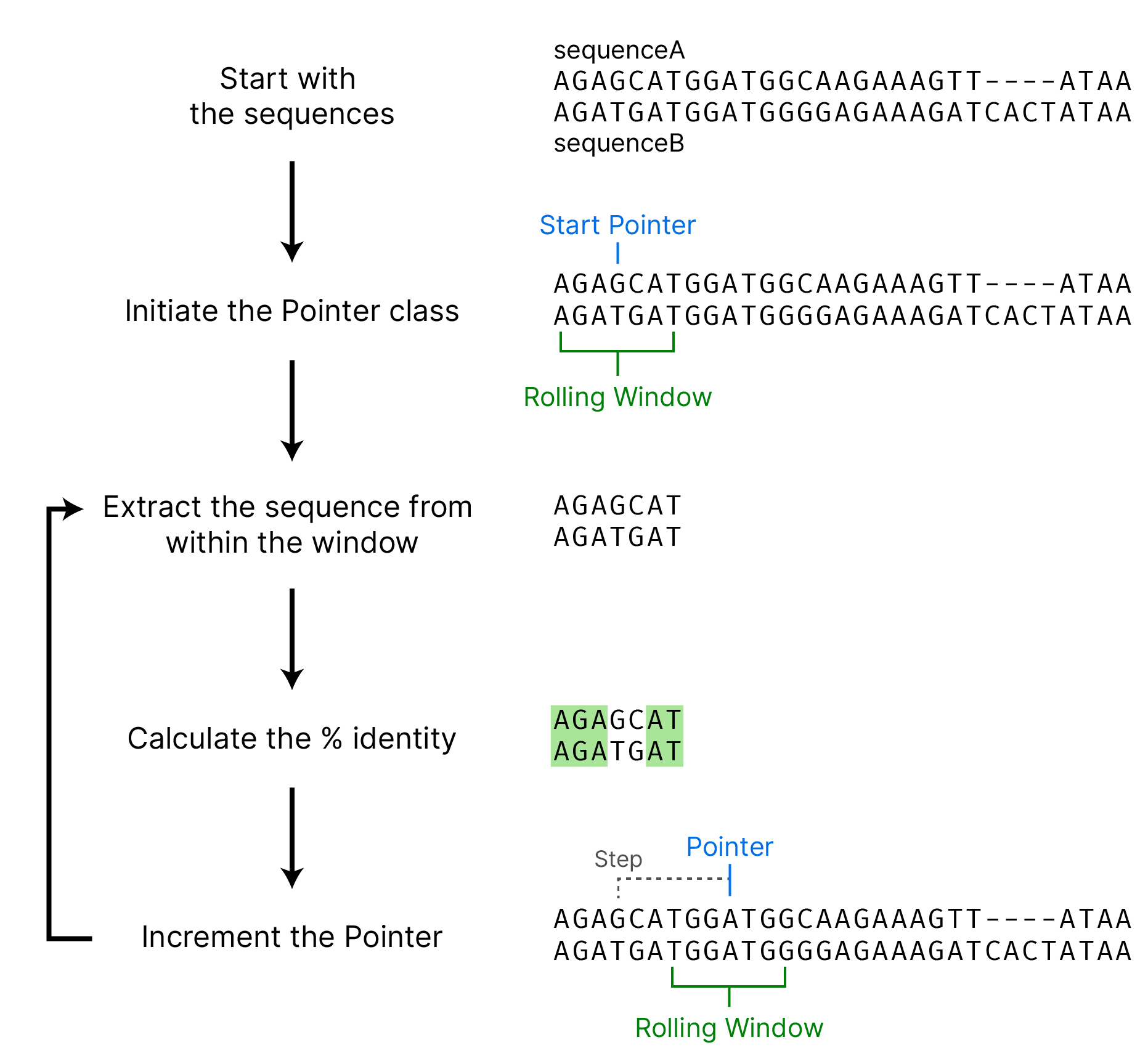
# Setup the basic params
window = 100
step = 5
# Initiate an empty dict to hold the identity values
combined_identities = {}
# Initiate the pointer class
pointer = Pointer(window,step)
# Iterate through the sequences
# but stop when the window goes past the last base/residue
while pointer.window_upper <= len(sequenceA):
# Extract the part of each sequence to compare
compareA = extract(pointer, sequenceA)
compareB = extract(pointer, sequenceA)
# Compare the sequences and get the % identity
identity = compare(compareA, compareB)
# Add the identity to the dict with the pointer as a key
combined_identities[pointer.pointer] = identity
# Increment the pointer and window location
pointer.increment()
# Complete! Let's see what the values are.
print(combined_identities)You’ll have noticed that we also don’t have a nice way to export the output yet. Ideally I would like to export a csv file, which can be read into something like Excel or R, but again this is past the scope of this post. I’ll include the functionality in the final script.
Playing with parameters
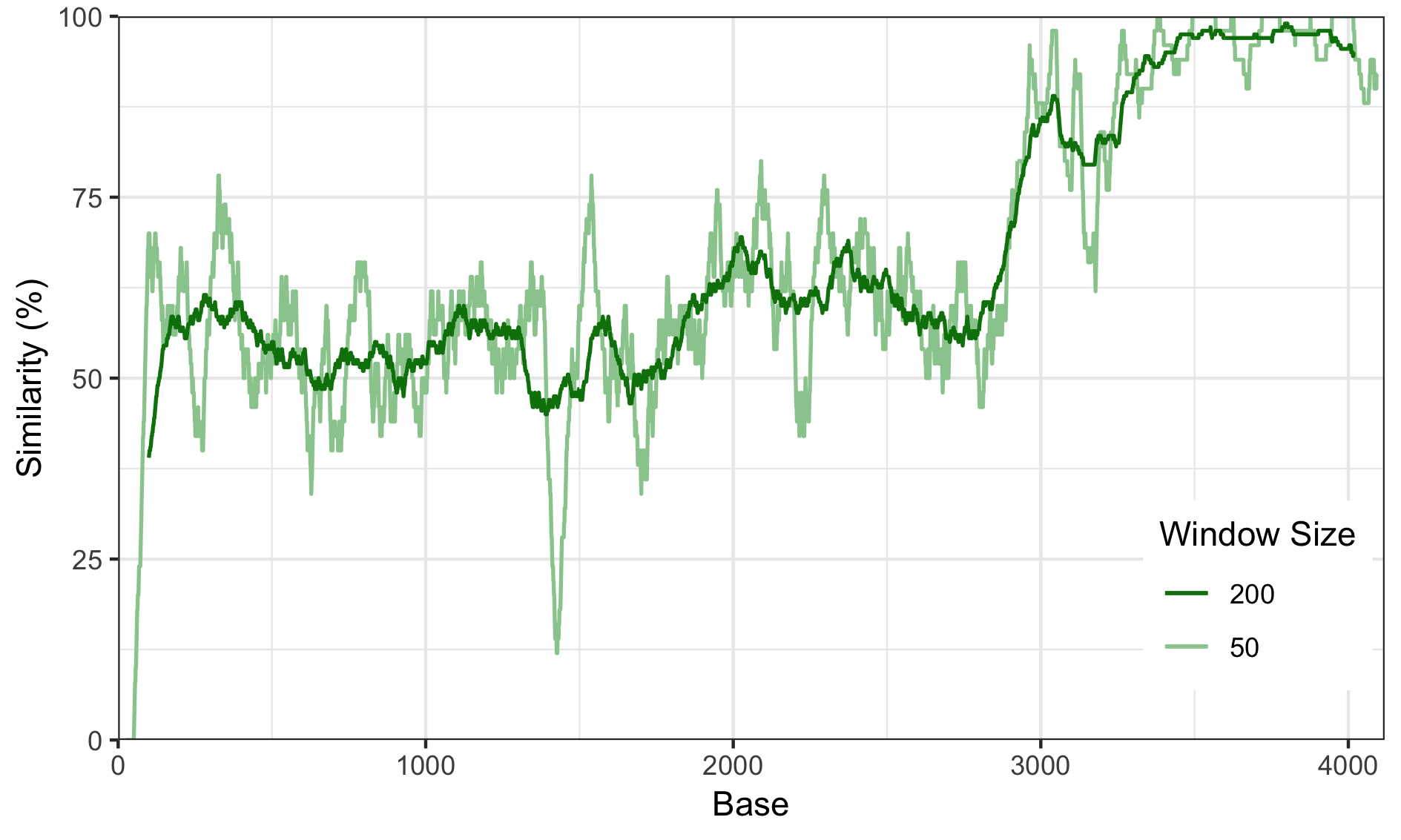
For the following plots, I took the output from above and make some nice figures using ggplot2 in R.
Adjusting the Window Size
Adjusting the window size has quite a dramatic effect. Too small and the plot will be filled with noise, too large and you’ll smooth out the curve so much that it will mask any nice features.
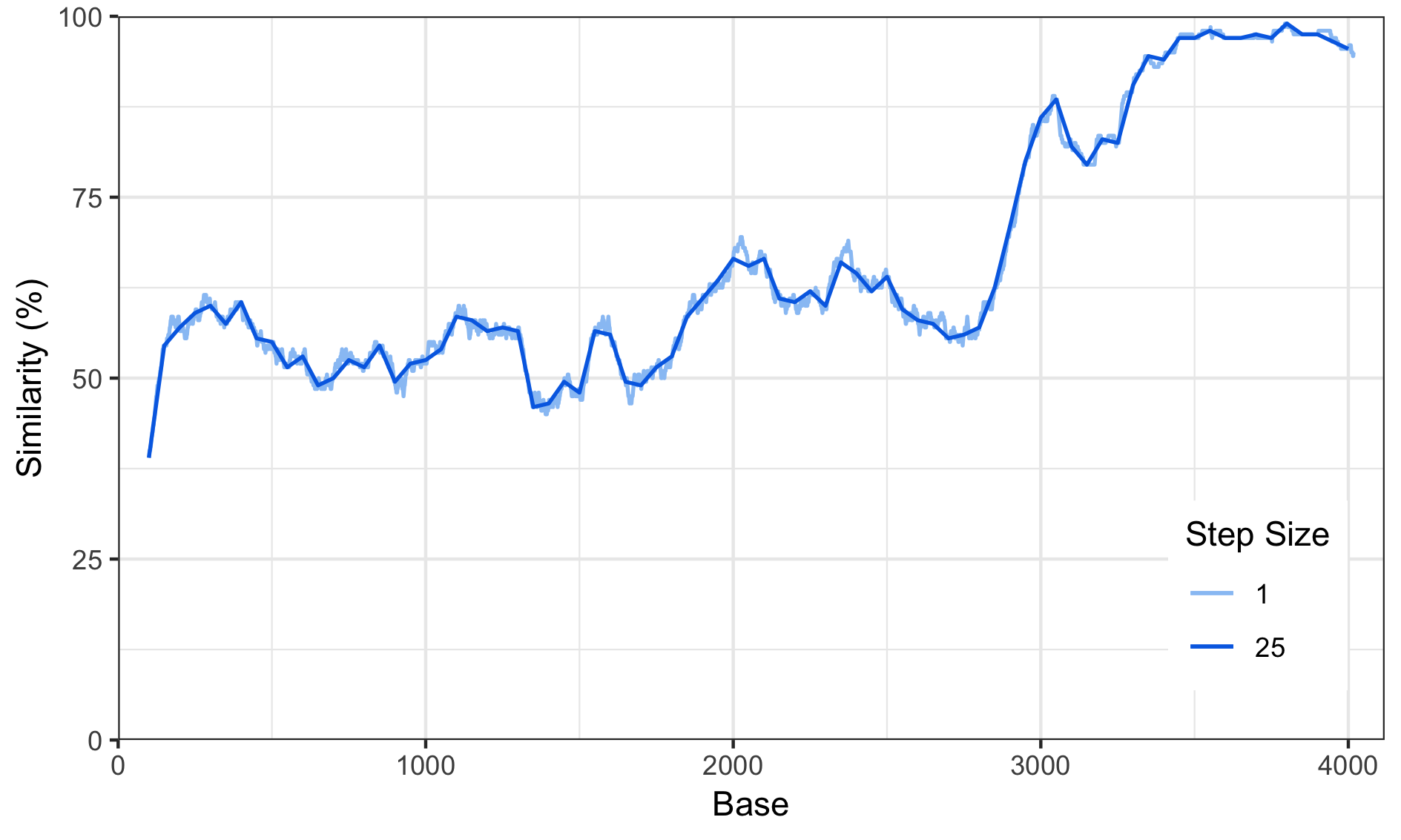
Adjusting the Step Size
Increasing the step size has a similiar effect as a low-pass filter, it will remove most of the high-frequency spikes.
Conclusions
Hopefully you’ve found this useful. To make the script useable, I’ve added some bits which enable you to read in alignments and export to CSV. As well as other snippets to make the code more robust. Find it on my GitHub and feel free to use, modify, and share as you see fit!